It searches by simulation the sample size (given other factors, such as effect sizes) or effect size (given other factors, such as sample size) with power to detect an effect close to a target value.
Usage
x_from_power(
object,
x = arg_x_from_power(object, "x", arg_in = "call") %||% "n",
pop_es_name = arg_x_from_power(object, "pop_es_name", arg_in = "call"),
target_power = 0.8,
what = arg_x_from_power(object, "what") %||% "point",
goal = arg_x_from_power(object, "goal") %||% {
switch(what, point = "ci_hit", ub
= "close_enough", lb = "close_enough")
},
ci_level = 0.95,
tolerance = NULL,
x_interval = switch(x, n = c(50, 2000), es = NULL),
extendInt = NULL,
progress = TRUE,
simulation_progress = NULL,
max_trials = NULL,
final_nrep = attr(object, "args")$nrep %||% (object$nrep_final %||% 400),
final_R = attr(object, "args")$R %||% (object$args$final_R %||% 1000),
seed = NULL,
x_include_interval = FALSE,
check_es_interval = TRUE,
power_curve_args = list(power_model = NULL, start = NULL, lower_bound = NULL,
upper_bound = NULL, nls_control = list(), nls_args = list()),
save_sim_all = FALSE,
algorithm = NULL,
control = list(),
internal_args = list(),
rejection_rates_args = list(collapse = "all_sig", at_least_k = 1, p_adjust_method =
"none", alpha = 0.05)
)
n_from_power(
object,
pop_es_name = NULL,
target_power = 0.8,
what = formals(x_from_power)$what,
goal = formals(x_from_power)$goal,
ci_level = 0.95,
tolerance = NULL,
x_interval = c(50, 2000),
extendInt = NULL,
progress = TRUE,
simulation_progress = NULL,
max_trials = NULL,
final_nrep = formals(x_from_power)$final_nrep,
final_R = formals(x_from_power)$final_R,
seed = NULL,
x_include_interval = FALSE,
check_es_interval = TRUE,
power_curve_args = list(power_model = NULL, start = NULL, lower_bound = NULL,
upper_bound = NULL, nls_control = list(), nls_args = list()),
save_sim_all = FALSE,
algorithm = NULL,
control = list(),
internal_args = list(),
rejection_rates_args = list(collapse = "all_sig", at_least_k = 1, p_adjust_method =
"none", alpha = 0.05)
)
n_region_from_power(
object,
pop_es_name = NULL,
target_power = 0.8,
ci_level = 0.95,
tolerance = NULL,
x_interval = c(50, 2000),
extendInt = NULL,
progress = TRUE,
simulation_progress = NULL,
max_trials = NULL,
final_nrep = formals(x_from_power)$final_nrep,
final_R = formals(x_from_power)$final_R,
seed = NULL,
x_include_interval = FALSE,
check_es_interval = TRUE,
power_curve_args = list(power_model = NULL, start = NULL, lower_bound = NULL,
upper_bound = NULL, nls_control = list(), nls_args = list()),
save_sim_all = FALSE,
algorithm = NULL,
control = list(),
internal_args = list(),
rejection_rates_args = list(collapse = "all_sig", at_least_k = 1, p_adjust_method =
"none", alpha = 0.05)
)
# S3 method for class 'x_from_power'
print(x, digits = 3, call = TRUE, ...)
# S3 method for class 'n_region_from_power'
print(x, digits = 3, ...)
arg_x_from_power(object, arg, arg_in = NULL)
pba_diagnosis(
a_out,
p_interval = c(0.05, 0.95),
posterior_xlim = c(0.01, 0.99),
which = NULL
)Arguments
- object
A
power4testobject, which is the output ofpower4test(). Can also be apower4test_by_nobject, the output ofpower4test_by_n(), or apower4test_by_esobject, the output ofpower4test_by_es(). For these two types of objects, the attempt with power closest to thetarget_powerwill be used asobject, and all other attempts in them will be included in the estimation of subsequent attempts and the final output. Last, it can also be the output of a previous call tox_from_power(), and the stored trials will be retrieved.- x
For
x_from_power(),xset the value to be searched. Can be"n", the sample size, or"es", the population value of a parameter (set bypop_es_name). For theprintmethod ofx_from_powerobjects, this is the output ofx_from_power().- pop_es_name
The name of the parameter. Required if
xis"es". See the help page ofptable_pop()on the names for the argumentpop_es.- target_power
The target power, a value greater than 0 and less than one.
- what
The value for which is searched: the estimate power (
"point"), the upper bound of the confidence interval ("ub"), or the lower bound of the confidence interval ("lb").- goal
The goal of the search. If
"ci_hit", then the goal is to find a value ofxwith the confidence interval of the estimated power including the target power. If"close_enough", then the goal is to find a value ofxwith the value inwhat"close enough" to the target power, defined by having an absolute difference with the target power less thantolerance.- ci_level
The level of confidence of the confidence intervals computed for the estimated power. Default is .95, denoting 95%.
- tolerance
Used when the goal is
"close_enough". IfNULL, set automatically based on the algorithm used.- x_interval
A vector of two values, the minimum value and the maximum values of
x, in the search for the values (sample sizes or population values). IfNULL, default whenx = "es", it will be determined internally.- extendInt
Whether
x_intervalcan be expanded when estimating the the values to try. The value will be passed to the argument of the same name instats::uniroot(). Ifxis"n", then the default value is"upX". That is, a value higher than the maximum inx_intervalis allowed, if predicted by the tentative model. Otherwise, the default value is"no". See the help page ofstats::uniroot()for further information.- progress
Logical. Whether the searching progress is reported.
- simulation_progress
Logical. Whether the progress in each call to
power4test(),power4test_by_n(), orpower4test_by_es()is shown. To be passed to theprogressargument of these functions. IfNULL, set automatically based on the algorithm used.- max_trials
The maximum number of trials in searching the value with the target power. Rounded up if not an integer. If
NULL, set automatically based on the algorithm used.- final_nrep
The number of replications in the final stage, also the maximum number of replications in each call to
power4test(),power4test_by_n(), orpower4test_by_es(). Ifobjectis an output ofpower4test()orx_from_power()and this argument is not set,final_nrepwill be set tonreporfinal_nrepstored inobject.- final_R
The number of Monte Carlo simulation or bootstrapping samples in the final stage. The
Rin callingpower4test(),power4test_by_n(), orpower4test_by_es()will be stepped up to this value when approaching the target power. Do not need to be very large because the goal is to estimate power by replications, not for high precision in one single replication. Ifobjectis an output ofpower4test()orx_from_power()and this argument is not set,final_Rwill be set toRorfinal_Rstored inobject.- seed
If not
NULL,set.seed()will be used to make the process reproducible. This is not always possible if many stages of parallel processing is involved.- x_include_interval
Logical. Whether the minimum and maximum values in
x_intervalare mandatory to be included in the values to be searched.- check_es_interval
If
TRUE, the default, andxis"es", a conservative probable range of valid values for the selected parameter will be determined, and it will be used instead ofx_interval. If the range spans both positive and negative values, only the interval of the same sign as the population value inobjectwill be used.- power_curve_args
A named list of arguments to be passed
power_curve()when estimating the relation between power andx(sample size or effect size). Please refer topower_curve()on available arguments. There is one except:power_modelis mapped to theformulaargument ofpower_curve().- save_sim_all
If
FALSE, the default, the data in eachpower4testobject for each value ofxis not saved, to reduce the size of the output. If set toTRUE, the size of the output can be very large in size.- algorithm
The algorithm for finding
x. Can be"power_curve","bisection", or"probabilistic_bisection". The default algorithm depends onx.- control
A named list of additional arguments to be passed to the algorithm to be used. For advanced users.
- internal_args
A named list of internal arguments. For internal testing. Do not use it.
- rejection_rates_args
Argument values to be used when
rejection_rates()is called, used to decide how rejection rates will be estimated. Only one single test is supported byx_from_power(). Therefore,merge_all_testsis alwaysTRUEand cannot be changed. The argumentcollapsecan be"all_sig","at_least_one_sig", or"at_least_k_sig"(it cannot be"none"). Please refer torejection_rates()for other possible arguments. These values, if set, will overwrite any stored settings inobject.- digits
The number of digits after the decimal when printing the results.
- call
Logical. Whether the call is printed.
- ...
Optional arguments. Not used for now.
- arg
The name of element to retrieve.
- arg_in
The name of the element from which an element is to be retrieved.
- a_out
The output of
x_from_power()and friends. The algorithm used must be"probabilistic_bisection".- p_interval
The range of the plot that "zooms" around the solution (or the median of in the final posterior probability distribution), expressed in terms of the area of the distribution.
- posterior_xlim
The range of the first plot of search history and the plot of posterior probability distribution, expressed in terms of the area of the distribution.
- which
If
a_outis a list with more than one output ofx_from_power(), such as the output ofn_region_from_power(),whichmust be set to the name of one such output ("below"or"above"for the output ofn_region_from_power(), or the output ofq_power_mediation_*).
Value
The function x_from_power()
returns an x_from_power object,
which is a list with the following
elements:
power4test_trials: The output ofpower4test_by_n()for all sample sizes examined, or ofpower4test_by_es()for all population values of the selected parameter examined.rejection_rates: The output ofrejection_rates().x_tried: The sample sizes or population values examined.power_tried: The estimated rejection rates for all the values examined.x_final: The sample size or population value in the solution.NAif a solution not found.power_final: The estimated power of the value in the solution.NAif a solution not found.i_final: The position of the solution inpower4test_trials.NAif a solution not found.ci_final: The confidence interval of the estimated power in the solution. The method is determined by the optionpower4mome.ci_method. IfNULLor"wilson", Wilson's (1927) method is used. If"norm", normal approximation is used.ci_level: The level of confidence ofci_final.nrep_final: The number of replications (nrep) when estimating the power in the solution.power_curve: The output ofpower_curve()when estimating the power curve.target_power: The requested target power.power_tolerance: The allowed difference between the solution's estimated power and the target power. Determined by the number of replications and the level of confidence of the confidence intervals.x_estimated: The value (sample size or population value) with the target power, estimated bypower_curve. This is used, when solution not found, to determine the range of the values to search when calling the function again.start: The time and date when the process started.end: The time and date when the process ended.time_spent: The time spent in doing the search.args: A named list of the arguments ofx_from_power()used in the search.call: The call when this function is called.
The function n_region_from_power()
returns a named list of two output of
n_from_power(), of the class
n_region_from_power. The output
with what = "ub" is named "below",
and the output with what = "lb" is
namd "above".
The print-method of x_from_power
objects returns the object x
invisibly.
It is called for its side effect.
The print-method of x_from_power_region
objects returns the object x
invisibly.
It is called for its side effect.
The function arg_x_from_power()
returns the requested argument if
available. If not available, it
returns NULL.
The function pba_diagnosis()
returns NULL invisibly. Called for
its side-effect.
Details
This is how to use x_from_power():
Specify the model by
power4test(), withdo_the_test = FALSE, and set the magnitude of the effect sizes to the minimum levels to detect.Add the test using
power4test()usingtest_funandtest_args(see the help page ofpower4test()for details). Run it on the starting sample size or effect size.Call
x_from_power()on the output ofpower4test()returned from the previous step. This function will iteratively repeat the analysis on either other sample sizes, or other values for a selected model parameter (the effect sizes), trying to achieve a goal (goal) for a value of interest (what).
If the goal is "ci_hit", the
search will try to find a value (a sample
size, or a population value of
the selected model parameter) with
a power level close enough to the
target power, defined by having its
confidence interval for the power
including the target power.
If the goal is "close_enough",
then the search will try to find a
value of x with its level of
power ("point"), the upper bound
of the confidence interval for this
level of power ("ub"), or the
lower bound of the confidence interval
fro this level of power ("lb")
"close enough" to the target level of
power, defined by having an absolute
difference less than the tolerance.
If several values of x (sample
size or the population value of
a model parameter) have already been
examined by power4test_by_n() or
power4test_by_es(), the output
of these two functions can also be
used as object by x_from_power().
Usually, the default values of the arguments should be sufficient.
The results can be viewed using
summary(), and the output has
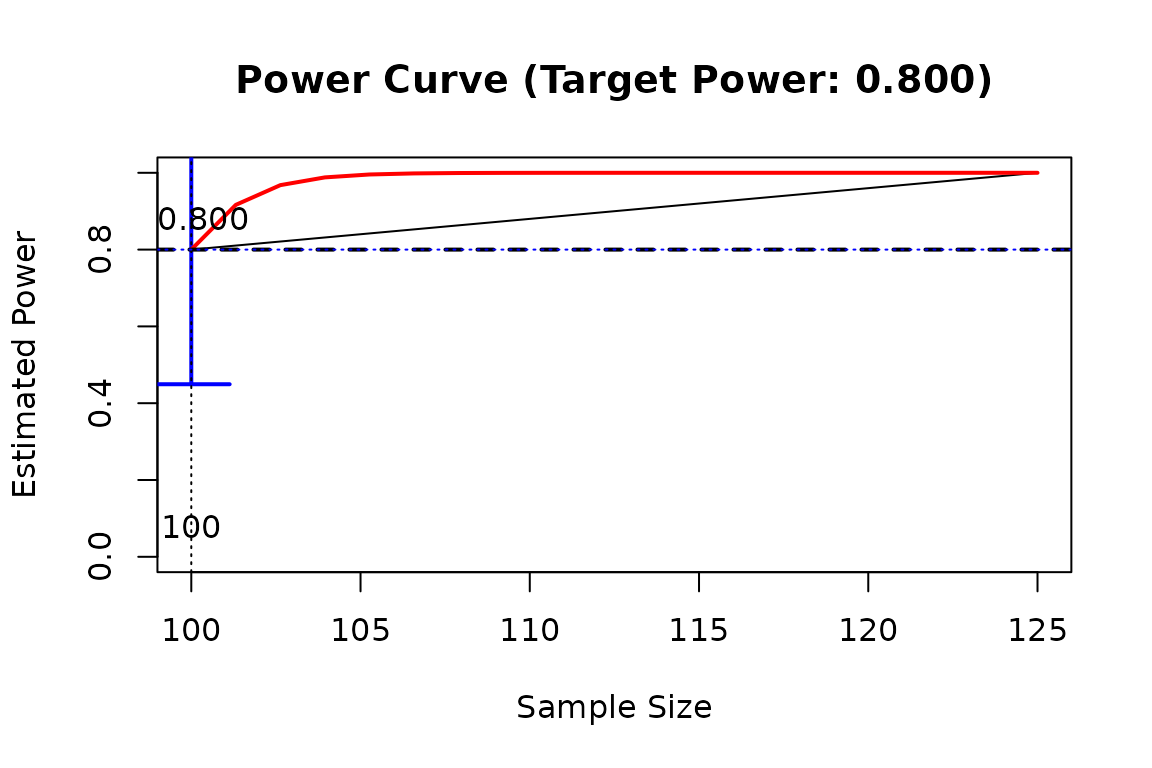
a plot method (plot.x_from_power()) to
plot the relation between power and
values (of x) examined.
A detailed illustration on how to use this function for sample size can be found from this page:
https://sfcheung.github.io/power4mome/articles/x_from_power_for_n.html
The function n_from_power() is just
a wrapper of x_from_power(), with
x set to "n".
The function n_region_from_power() is just
a wrapper of x_from_power(), with
x set to "n", with two passes, one
with what = "ub" and one with
what = "lb".
The print method only prints
basic information. Call the
summary method of x_from_power objects
(summary.x_from_power()) and its
print method for detailed results
The function arg_x_from_power()
is a helper to set argument values
if object is an output
of x_from_power() or similar
functions.
The function pba_diagnosis()
generates simple diagnostic plots
for the search history of probabilistic
bisection algorithm. This function is for advanced users
to examine the search history of
the probabilistic bisection algorithm.
It is for diagnostic purpose and has
limited support for customizing the plots.
Algorithms
Three algorithms are currently available, the simple (though sometimes inefficient) bisection method, a method that makes use of the estimated crude power curve, and probabilistic bisection algorithm (Waeber et al., 2013; see Chalmers, 2024, for applying this algorithm to power analysis).
Unlike typical root-finding problems,
the prediction of the level of power
is stochastic. Moreover, the computational
cost is high when Monte Carlo or
bootstrap confidence intervals are
used to do a test because the estimation
of the power for one single value of
x can sometimes take one minute or
longer. Therefore, in addition to
the simple bisection method, two methods,
named power curve method
(which belongs to the family of
surrogate function approximation method
reviewed in Chalmers, 2024) and
probabilistic bisection method
(Chalmers, 2024; Waeber et al., 2013) were also
developed for this
scenario.
Bisection Method
This method (called informat bisection
in Chalmers, 2024), enabled by
algorithm = "bisection",
basically starts with
an interval that probably encloses the
value of x that meets the goal,
and then successively narrows this
interval. A point inside this
interval, usually the mid-point but
can also be approximated by another
method (e.g., the power curve method),
is used as the estimate.
Though simple, there are cases in
which it can be slow. Nevertheless,
preliminary examination suggests that
this method is good enough for
scenarios in which only an approximate
sample size or range of sample sizes is
needed.
The internal workflow of
this method implemented in
x_from_power() will be presented
in a forthcoming technical vignette.
Power Curve Method
This method (belongs to the
surrogate function approximation family
reviewed in Chalmers, 2024),
enabled by algorithm = "power_curve",
starts with a crude
power curve based on a few points.
This tentative model is then used
to suggest the values to examine in
the next iteration. Unlike some other
implementations
of this family of methods, the form, not
just the parameters, of the
model can change across iterations,
as more and more data points are
available.
This method is the default method
for some scenarios, such as
x = "es" with goal = "ci_hit"
because the relation
between the power and the population
value of a parameter varies across
parameters, unlike the relation
between power and sample size, which
is monotonic. Therefore,
taking into account the working
power curve may help finding the
desired value of x.
Before version 0.1.1.33, this
method can be used only with
the goal "ci_hit". Since
version 0.1.1.34, it supports all
goals, like the bisection method.
The internal workflow of
this method implemented in
x_from_power() can be found in
this technical vignette: https://sfcheung.github.io/power4mome/articles/x_from_power_workflow.html.
Probabilistic Bisection
This method, proposed by Waeber and others (2013) for stochastic root-solving, has been adapted by Chalmers (2024) for power analysis. Similar to bisection, this method starts with an interval, with a initial probability for each value (or range of values), such as sample sizes, as the sample size with the target power. A value is then selected to estimate power (the median, by default). Based on the estimated power for this value, the distribution of probabilities is updated. This process is repeated until some termination criteria are met.
Unlike bisection, each iteration can be conducted with a small number of replications (e.g., 50). The method accumulate evidence in a Bayesian approach, and so the certainty of the solution is based on the accumulation of evidence from successive iterations, not on having strong evidence from a few iterations.
In x_from_power, the version
of probabilistic bisection proposed
by Waeber et al. (2013) was implemented,
with minor changes.
Most importantly, when some termination
criteria are met, the candidate value
of x (e.g., a sample size) will be
checked using a larger number of
replications (set by final_nrep)
to ensure that the estimated power
is indeed close enough to the target
power (based on tolerance value,
determined internally but can also be
set directly). If yes, it will be
returned as the solution. If no, then
the search will continue, until a
maximum number of candidate values
has been checked.
Although the bisection method can be fast in some situations (e.g., when the interval is narrow and the solution happens to be inside the interval), the probabilistic bisection method is nearly guaranteed to converge to the solution (as long as the solution is inside the interval). Therefore, this is the default algorithm in some scenarios.
The internal workflow of
this method implemented in
x_from_power() will be presented
in a forthcoming technical vignette.
References
Chalmers, R. P. (2024). Solving variables with Monte Carlo simulation experiments: A stochastic root-solving approach. Psychological Methods. Advance online publication. doi:10.1037/met0000689
Waeber, R., Frazier, P. I., & Henderson, S. G. (2013). Bisection search with noisy responses. SIAM Journal on Control and Optimization, 51(3), 2261–2279. doi:10.1137/120861898
Wilson, E. B. (1927). Probable inference, the law of succession, and statistical inference. Journal of the American Statistical Association, 22(158), 209-212. doi:10.1080/01621459.1927.10502953
Examples
# Specify the population model
mod <-
"
m ~ x
y ~ m + x
"
# Specify the population values
mod_es <-
"
m ~ x: m
y ~ m: l
y ~ x: n
"
# Generate the datasets
sim_only <- power4test(nrep = 5,
model = mod,
pop_es = mod_es,
n = 100,
do_the_test = FALSE,
iseed = 2345)
#> Simulate the data:
#> Fit the model(s):
# Do a test
test_out <- power4test(object = sim_only,
test_fun = test_parameters,
test_args = list(pars = "m~x"))
#> Do the test: test_parameters: CIs (pars: m~x)
# Determine the sample size with a power of .80 (default)
# In real analysis, to have more stable results:
# - Use a larger final_nrep (e.g., 400).
# If the default values are OK, this call is sufficient:
# power_vs_n <- x_from_power(test_out,
# x = "n",
# seed = 4567)
power_vs_n <- x_from_power(test_out,
x = "n",
progress = TRUE,
target_power = .80,
final_nrep = 5,
max_trials = 1,
seed = 1234)
#>
#> --- Setting ---
#>
#> Algorithm: bisection
#> Goal: ci_hit
#> What: point (Estimated Power)
#>
#> --- Progress ---
#>
#> - Set 'progress = FALSE' to suppress displaying the progress.
#> - Set 'simulation progress = FALSE' to suppress displaying the progress
#> in the simulation.
#>
#> Initial interval: [50, 100]
#>
#>
#> Do the simulation for the lower bound:
#>
#> Try x = 50
#>
#> Updating the simulation for sample size: 50
#> Re-simulate the data:
#> Fit the model(s):
#> Update the test(s):
#> Update test_parameters: CIs (pars: m~x) :
#>
#> Estimated power at 50: 0.400, 95.0% confidence interval: [0.118,0.769]
#>
#> Initial interval: [50, 100]
#>
#> - Rejection Rates:
#> [test]: test_parameters: CIs (pars: m~x)
#> [test_label]: m~x
#> n est p.v reject r.cilo r.cihi
#> 1 50 0.222 1.000 0.400 0.118 0.769
#> 2 100 0.298 1.000 0.800 0.376 0.964
#>
#>
#> == Enter extending interval ...
#> The interval is already valid: [50, 100]
#> == Exit extending interval ...
#>
#> Iteration # 1
#>
#> Try x = 75
#>
#> Updating the simulation for sample size: 75
#> Re-simulate the data:
#> Fit the model(s):
#> Update the test(s):
#> Update test_parameters: CIs (pars: m~x) :
#>
#> Estimated power at 75: 1.000, 95.0% confidence interval: [0.566,1.000]
#> - Rejection Rates:
#> [test]: test_parameters: CIs (pars: m~x)
#> [test_label]: m~x
#> n est p.v reject r.cilo r.cihi
#> 1 50 0.222 1.000 0.400 0.118 0.769
#> 2 75 0.334 1.000 1.000 0.566 1.000
#> 3 100 0.298 1.000 0.800 0.376 0.964
#>
#> - 'nls()' estimation skipped when less than 4 values of predictor examined.
#> Solution found.
#>
#> ========== Final Stage ==========
#>
#> - Start at 2026-03-08 15:33:00
#> - Rejection Rates:
#>
#> [test]: test_parameters: CIs (pars: m~x)
#> [test_label]: m~x
#> n est p.v reject r.cilo r.cihi
#> 1 50 0.222 1.000 0.400 0.118 0.769
#> 2 75 0.334 1.000 1.000 0.566 1.000
#> 3 100 0.298 1.000 0.800 0.376 0.964
#> Notes:
#> - n: The sample size in a trial.
#> - p.v: The proportion of valid replications.
#> - est: The mean of the estimates in a test across replications.
#> - reject: The proportion of 'significant' replications, that is, the
#> rejection rate. If the null hypothesis is true, this is the Type I
#> error rate. If the null hypothesis is false, this is the power.
#> - r.cilo,r.cihi: The confidence interval of the rejection rate, based
#> on Wilson's (1927) method.
#> - Refer to the tests for the meanings of other columns.
#>
#> - Estimated Power Curve:
#>
#> Call:
#> power_curve(object = by_x_1, formula = power_model, start = power_curve_start,
#> lower_bound = lower_bound, upper_bound = upper_bound, nls_args = nls_args,
#> nls_control = nls_control, verbose = progress)
#>
#> Predictor: n (Sample Size)
#>
#> Model:
#>
#> Call: stats::glm(formula = reject ~ x, family = "binomial", data = reject1)
#>
#> Coefficients:
#> (Intercept) x
#> -2.25384 0.04624
#>
#> Degrees of Freedom: 14 Total (i.e. Null); 13 Residual
#> Null Deviance: 17.4
#> Residual Deviance: 15.23 AIC: 19.23
#>
#>
#> - Final Value: 75
#>
#> - Final Estimated Power: 1.0000
#> - Confidence Interval: [0.5655; 1.0000]
#> - CI Level: 95.00%
summary(power_vs_n)
#>
#> ====== x_from_power Results ======
#>
#> Call:
#> x_from_power(object = test_out, x = "n", target_power = 0.8,
#> progress = TRUE, max_trials = 1, final_nrep = 5, seed = 1234)
#>
#> Predictor (x): Sample Size
#>
#> - Target Power: 0.800
#> - Goal: Find 'x' with the confidence interval of the estimated power
#> enclosing the target power.
#>
#> === Major Results ===
#>
#> - Final Value (Sample Size): 75
#>
#> - Final Estimated Power: 1.000
#> - Confidence Interval: [0.566; 1.000]
#> - Level of confidence: 95.0%
#> - Based on 5 replications.
#>
#> === Technical Information ===
#>
#> - Algorithm: bisection
#> - The range of values explored: 50 to 100
#> - Time spent in the search: 0.9439 secs
#> - The final crude model for the power-predictor relation:
#>
#> Model Type: Logistic Regression
#>
#> Call:
#> power_curve(object = by_x_1, formula = power_model, start = power_curve_start,
#> lower_bound = lower_bound, upper_bound = upper_bound, nls_args = nls_args,
#> nls_control = nls_control, verbose = progress)
#>
#> Predictor: n (Sample Size)
#>
#> Model:
#>
#> Call: stats::glm(formula = reject ~ x, family = "binomial", data = reject1)
#>
#> Coefficients:
#> (Intercept) x
#> -2.25384 0.04624
#>
#> Degrees of Freedom: 14 Total (i.e. Null); 13 Residual
#> Null Deviance: 17.4
#> Residual Deviance: 15.23 AIC: 19.23
#>
#> - Detailed Results:
#>
#> [test]: test_parameters: CIs (pars: m~x)
#> [test_label]: m~x
#> n est p.v reject r.cilo r.cihi
#> 1 50 0.222 1.000 0.400 0.118 0.769
#> 2 75 0.334 1.000 1.000 0.566 1.000
#> 3 100 0.298 1.000 0.800 0.376 0.964
#> Notes:
#> - n: The sample size in a trial.
#> - p.v: The proportion of valid replications.
#> - est: The mean of the estimates in a test across replications.
#> - reject: The proportion of 'significant' replications, that is, the
#> rejection rate. If the null hypothesis is true, this is the Type I
#> error rate. If the null hypothesis is false, this is the power.
#> - r.cilo,r.cihi: The confidence interval of the rejection rate, based
#> on Wilson's (1927) method.
#> - Refer to the tests for the meanings of other columns.
#>
plot(power_vs_n)