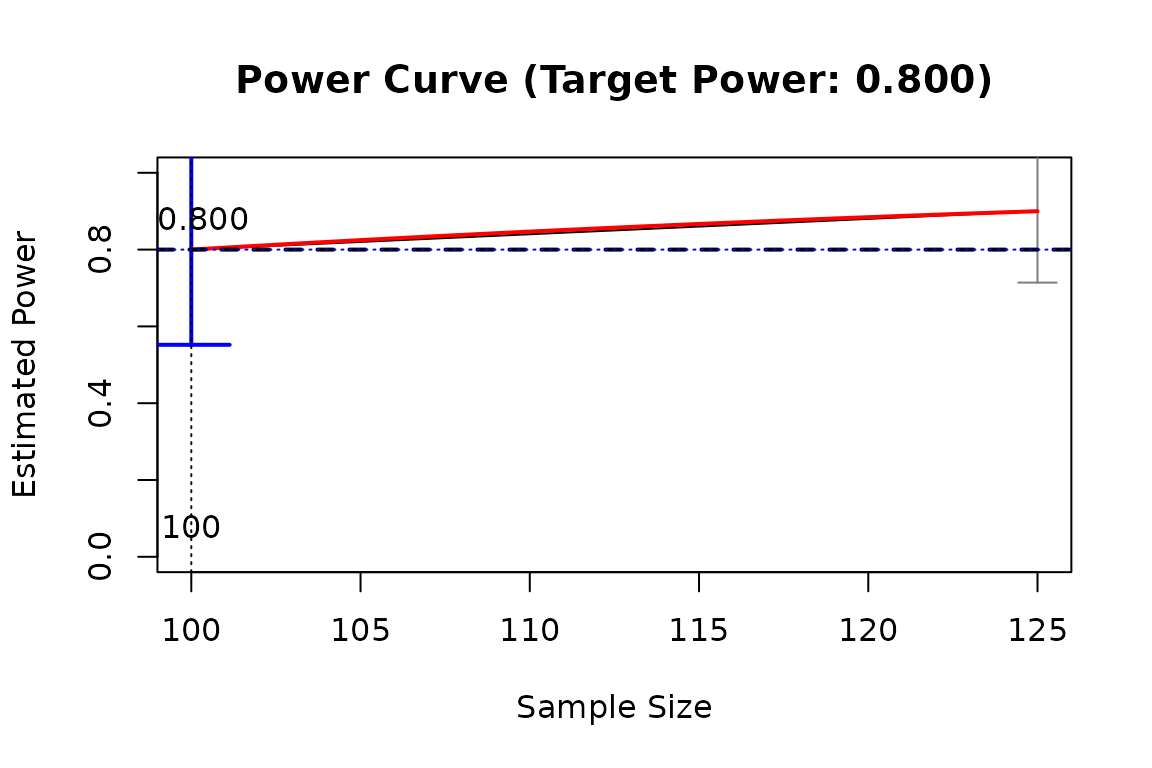
It plots the results of 'x_from_power', such as the estimated power against sample size.
Usage
# S3 method for class 'x_from_power'
plot(
x,
what = c("ci", "power_curve", "final_x", "final_power", "target_power", switch(x$x, n =
"sig_area", es = NULL)),
text_what = c("final_x", "final_power", switch(x$x, n = "sig_area", es = NULL)),
digits = 3,
main = NULL,
xlab = NULL,
ylab = "Estimated Power",
ci_level = 0.95,
pars_ci = list(),
pars_power_curve = list(),
pars_ci_final_x = list(lwd = 3, length = 0.2, col = "blue"),
pars_target_power = list(lty = "dashed", lwd = 2, col = "black"),
pars_final_x = list(lty = "dotted"),
pars_final_power = list(lty = "dotted", col = "blue"),
pars_text_final_x = list(y = 0, pos = 3, cex = 1),
pars_text_final_power = list(pos = 3, cex = 1),
pars_sig_area = list(col = adjustcolor("lightblue", alpha.f = 0.1)),
pars_text_sig_area = list(cex = 1),
prop_of_trials = NULL,
min_trials_for_prop = NULL,
override_for_pba = TRUE,
...
)
# S3 method for class 'n_region_from_power'
plot(
x,
what = c("ci", "power_curve", "final_x", "final_power", "target_power", "sig_area"),
text_what = c("final_x", "final_power", "sig_area"),
digits = 3,
main = paste0("Power Curve ", "(Target Power: ", formatC(x$below$target_power, digits =
digits, format = "f"), ")"),
xlab = NULL,
ylab = "Estimated Power",
ci_level = 0.95,
pars_ci = list(),
pars_power_curve = list(),
pars_ci_final_x = list(lwd = 2, length = 0.2, col = "blue"),
pars_target_power = list(lty = "dashed", lwd = 2, col = "black"),
pars_final_x = list(lty = "dotted"),
pars_final_power = list(lty = "dotted", col = "blue"),
pars_text_final_x = list(pos = 3, cex = 1),
pars_text_final_x_lower = pars_text_final_x,
pars_text_final_x_upper = pars_text_final_x,
pars_text_final_power = list(cex = 1),
pars_sig_area = list(col = adjustcolor("lightblue", alpha.f = 0.1)),
pars_text_sig_area = list(cex = 1),
...
)Arguments
- x
An
x_from_powerobject, the output ofx_from_power().- what
A character vector of what to include in the plot. Possible values are
"ci"(confidence intervals for the estimated value of the predictor),"power_curve"(the crude power curve, if available),"final_x"(a vertical line for the value of the predictor with estimated power close enough to the target power by confidence interval),"final_power"(a horizontal line for the estimated power of the final value of the predictor),"target_power"(a horizontal line for the target power), and"sig_area"(the area significantly higher or lower than the target power, ifgoalis"close_enough"andwhatis"lb"or"ub"). By default, all these elements will be plotted.- text_what
A character vector of what numbers to be added as labels. Possible values are
"final_x"(the value of the predictor with estimated power close enough to the target power by confidence interval)"final_power"(the estimated power of the final value of the predictor), and"sig_area"(labeling the area significantly higher or lower than the target power, ifgoalis"close_enough"andwhatis"lb"or"ub"). By default, all these labels will be added.- digits
The number of digits after the decimal that will be used when adding numbers.
- main
The title of the plot. If
NULL, it will be generated automatically.- xlab, ylab
The labels for the horizontal and vertical axes, respectively.
- ci_level
The level of confidence of the confidence intervals, if requested. Default is
.95, denoting 95%.- pars_ci
A named list of arguments to be passed to
arrows()to customize the drawing of the confidence intervals.- pars_power_curve
A named list of arguments to be passed to
points()to customize the drawing of the power curve.- pars_ci_final_x
A named list of arguments to be passed to
arrows()to customize the drawing of the confidence interval of the final value of the predictor.- pars_target_power
A named list of arguments to be passed to
abline()when drawing the horizontal line for the target power.- pars_final_x
A named list of arguments to be passed to
abline()when drawing the vertical line for the final value of the predictor.- pars_final_power
A named list of arguments to be passed to
abline()when drawing the horizontal line for the estimated power at the final value of the predictor.- pars_text_final_x
A named list of arguments to be passed to
text()when adding the label for the final value of the predictor.- pars_text_final_power
A named list of arguments to be passed to
text()when adding the label for the estimated power of final value of the predictor.- pars_sig_area
A named list of arguments to be passed to
rect()when shading the area significantly higher or lower than the target power.- pars_text_sig_area
A named list of arguments to be passed to
text()when labelling the area significantly higher or lower than the target power.- prop_of_trials
The proportion of trials to be included in the plot. If
NULL, it will be determined based on the algorithm used.- min_trials_for_prop
The minimum number of trials for
prop_of_trialsto be used. IfNULL, it will be determined based on the algorithm used.- override_for_pba
If
TRUE, the default, the values of some arguments will be overriden internally if the algorithm used is"probabilistic_bisection", to make the plot suitable for this algorithm. For example, the power curve and the confidence intervals for trials other than the solution will not be plotted, even if requested. IfFALSE, then argument values will be not be changed internally.- ...
Optional arguments. Passed to
plot()when drawing the estimated power against the predictor.- pars_text_final_x_lower, pars_text_final_x_upper
If two values of the predictor are to be printed, these are the named list of the arguments to be passed to
text()when adding the labels for these two values.
Value
The plot-method of x_from_power
returns x invisibly.
It is called for its side effect.
The plot-method of n_region_from_power
returns x invisibly.
It is called for its side effect.
Details
The plot method of x_from_power
objects currently plots the relation
between estimated power and
the values examined by x_from_power().
Other elements
can be requested (see the argument
what), and they can be customized
individually.
The plot-method for
n_region_from_power objects is
a modified version of the plot-method
for x_from_power. It plots the
results of two runs of n_from_power()
in one plot. It is otherwise similar
to the plot-method for x_from_power.
Examples
# Specify the population model
mod <-
"
m ~ x
y ~ m + x
"
# Specify the population values
mod_es <-
"
m ~ x: m
y ~ m: l
y ~ x: n
"
# Generate the datasets
sim_only <- power4test(nrep = 10,
model = mod,
pop_es = mod_es,
n = 100,
do_the_test = FALSE,
iseed = 1234)
#> Simulate the data:
#> Fit the model(s):
# Do a test
test_out <- power4test(object = sim_only,
test_fun = test_parameters,
test_args = list(pars = "m~x"))
#> Do the test: test_parameters: CIs (pars: m~x)
# Determine the sample size with a power of .80 (default)
power_vs_n <- x_from_power(test_out,
x = "n",
progress = TRUE,
target_power = .80,
final_nrep = 10,
max_trials = 1,
seed = 2345)
#>
#> --- Setting ---
#>
#> Algorithm: bisection
#> Goal: ci_hit
#> What: point (Estimated Power)
#>
#> --- Progress ---
#>
#> - Set 'progress = FALSE' to suppress displaying the progress.
#> - Set 'simulation progress = FALSE' to suppress displaying the progress
#> in the simulation.
#>
#> Initial interval: [50, 100]
#>
#>
#> Do the simulation for the lower bound:
#>
#> Try x = 50
#>
#> Updating the simulation for sample size: 50
#> Re-simulate the data:
#> Fit the model(s):
#> Update the test(s):
#> Update test_parameters: CIs (pars: m~x) :
#>
#> Estimated power at 50: 0.700, 95.0% confidence interval: [0.397,0.892]
#>
#> Initial interval: [50, 100]
#>
#> - Rejection Rates:
#> [test]: test_parameters: CIs (pars: m~x)
#> [test_label]: m~x
#> n est p.v reject r.cilo r.cihi
#> 1 50 0.307 1.000 0.700 0.397 0.892
#> 2 100 0.320 1.000 0.800 0.490 0.943
#>
#> One of the bounds in the interval is already a solution.
#>
#> - 'nls()' estimation skipped when less than 4 values of predictor examined.
#> Solution found.
#>
#> ========== Final Stage ==========
#>
#> - Start at 2026-03-08 15:32:14
#> - Rejection Rates:
#>
#> [test]: test_parameters: CIs (pars: m~x)
#> [test_label]: m~x
#> n est p.v reject r.cilo r.cihi
#> 1 50 0.307 1.000 0.700 0.397 0.892
#> 2 100 0.320 1.000 0.800 0.490 0.943
#> Notes:
#> - n: The sample size in a trial.
#> - p.v: The proportion of valid replications.
#> - est: The mean of the estimates in a test across replications.
#> - reject: The proportion of 'significant' replications, that is, the
#> rejection rate. If the null hypothesis is true, this is the Type I
#> error rate. If the null hypothesis is false, this is the power.
#> - r.cilo,r.cihi: The confidence interval of the rejection rate, based
#> on Wilson's (1927) method.
#> - Refer to the tests for the meanings of other columns.
#>
#> - Estimated Power Curve:
#>
#> Call:
#> power_curve(object = by_x_1, formula = power_model, start = power_curve_start,
#> lower_bound = lower_bound, upper_bound = upper_bound, nls_args = nls_args,
#> nls_control = nls_control, verbose = progress)
#>
#> Predictor: n (Sample Size)
#>
#> Model:
#>
#> Call: stats::glm(formula = reject ~ x, family = "binomial", data = reject1)
#>
#> Coefficients:
#> (Intercept) x
#> 0.30830 0.01078
#>
#> Degrees of Freedom: 19 Total (i.e. Null); 18 Residual
#> Null Deviance: 22.49
#> Residual Deviance: 22.23 AIC: 26.23
#>
#>
#> - Final Value: 50
#>
#> - Final Estimated Power: 0.7000
#> - Confidence Interval: [0.3968; 0.8922]
#> - CI Level: 95.00%
plot(power_vs_n)