Plot a network of models
generated by model_graph().
Usage
# S3 method for class 'model_graph'
plot(x, ...)Arguments
- x
The output of
model_graph(). (Namedxbecause it is required in the naming of arguments of theplotgeneric function.)- ...
Additional arguments, passed to the
plot-method of anigraphobject.
Details
This function is the plot
method of model_graph objects,
the output of
model_graph().
For now, it simply passes the object
to plot-method of an igraph object.
This function
is created for possible customization
of the plot in the future.
Examples
library(lavaan)
dat <- dat_path_model
mod <-
"
x3 ~ a*x1 + b*x2
x4 ~ a*x1
ab := a*b
"
fit <- sem(mod, dat_path_model, fixed.x = TRUE)
out <- model_set(fit)
#>
#> Generate 2 less restrictive model(s):
#>
| | 0 % ~calculating
|+++++++++++++++++++++++++ | 50% ~00s
|++++++++++++++++++++++++++++++++++++++++++++++++++| 100% elapsed=00s
#>
#> Generate 2 more restrictive model(s):
#>
| | 0 % ~calculating
|+++++++++++++++++++++++++ | 50% ~00s
|++++++++++++++++++++++++++++++++++++++++++++++++++| 100% elapsed=00s
#>
#> Check for duplicated models (5 model[s] to check):
#>
|
| | 0%
|
|+++++ | 10%
|
|++++++++++ | 20%
|
|+++++++++++++++ | 30%
|
|++++++++++++++++++++ | 40%
|
|+++++++++++++++++++++++++ | 50%
|
|++++++++++++++++++++++++++++++ | 60%
|
|+++++++++++++++++++++++++++++++++++ | 70%
|
|++++++++++++++++++++++++++++++++++++++++ | 80%
|
|+++++++++++++++++++++++++++++++++++++++++++++ | 90%
|
|++++++++++++++++++++++++++++++++++++++++++++++++++| 100%
#>
#> Fit the 5 model(s) (duplicated models removed):
out
#>
#> Call:
#> model_set(sem_out = fit)
#>
#> Number of model(s) fitted : 5
#> Number of model(s) converged : 5
#> Number of model(s) passed post.check: 5
#>
#> The models (sorted by BPP):
#> model_df df_diff Prior BIC BPP cfi rmsea srmr
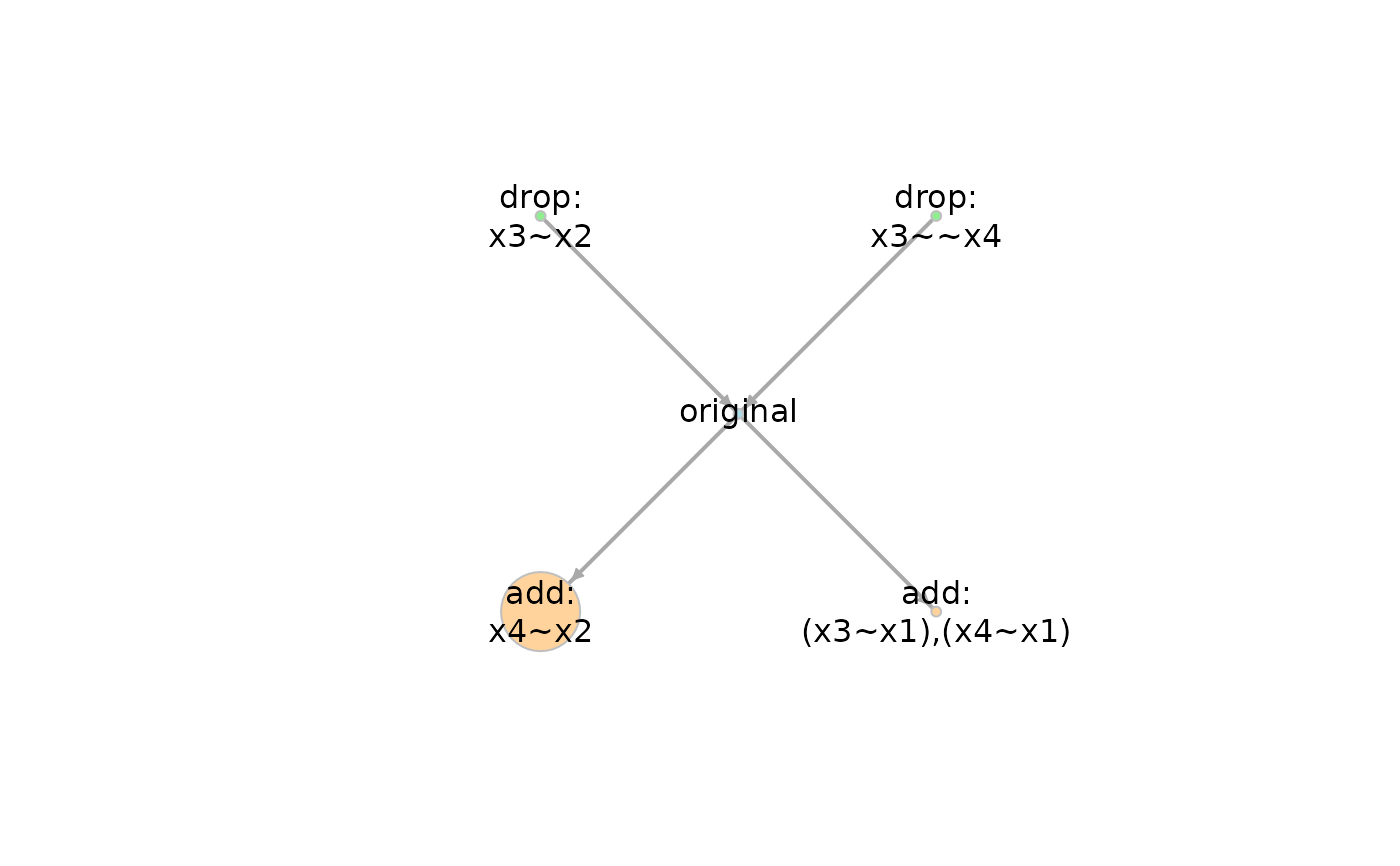
#> add: x4~x2 1 1 0.200 400.291 1.000 1.000 0.017 0.023
#> original 2 0 0.200 431.452 0.000 0.736 0.417 0.194
#> add: (x3~x1),(x4~x1) 1 1 0.200 435.397 0.000 0.733 0.593 0.193
#> drop: x3~~x4 3 -1 0.200 441.229 0.000 0.634 0.401 0.231
#> drop: x3~x2 3 -1 0.200 455.926 0.000 0.522 0.458 0.255
#>
#> Note:
#> - BIC: Bayesian Information Criterion.
#> - BPP: BIC posterior probability.
#> - model_df: Model degrees of freedom.
#> - df_diff: Difference in df compared to the original/target model.
#> - To show cumulative BPPs, call print() with 'cumulative_bpp = TRUE'.
#> - At least one model has fixed.x = TRUE. The models are not checked for
#> equivalence.
#> - Since Version 0.1.3.5, the default values of exclude_feedback and
#> exclude_xy_cov changed to TRUE. Set them to FALSE to reproduce
#> results from previous versions.
g <- model_graph(out)
plot(g)