Set the colors of selected edges.
Arguments
- semPaths_plot
A qgraph::qgraph object generated by semPlot::semPaths, or a similar qgraph object modified by other semptools functions.
- color_list
A named vector or a list of named list. See the Details section on how to set this argument.
Value
A qgraph::qgraph based on the original one, with colors for selected edges changed.
Details
Modified a qgraph::qgraph object generated by semPlot::semPaths and change the colors of selected edges.
Setting the value of color_list
This argument can be set in two ways.
For a named vector, the name of an
element should be the path as
specified by lavaan::model.syntax
or as appeared in
lavaan::parameterEstimates().
For example, to change the color of the
path regressing y on x, the name
should be "y ~ x". To change the
color of the covariance between x1
and x2, the name should be "x1 ~~ x2". Therefore, c("y ~ x1" = "red", "x1 ~~ x2" = "blue") changes the
colors of the path from x1 to y
and the covariance between x1 and
x2 to "red" and "blue",
respectively.
The order of the two nodes may matter for covariances. Therefore, if the attribute of a covariance is not changed, try switching the order of the two nodes.
For a list of named lists, each named
list should have three named values:
from, to, and new_color. The
attribute of the edge from from to
to will be set to new_color.
The second approach is no longer recommended, though kept for backward compatibility.
Examples
mod_pa <-
'x1 ~~ x2
x3 ~ x1 + x2
x4 ~ x1 + x3
'
fit_pa <- lavaan::sem(mod_pa, pa_example)
lavaan::parameterEstimates(fit_pa)[, c("lhs", "op", "rhs", "est", "pvalue")]
#> lhs op rhs est pvalue
#> 1 x1 ~~ x2 0.005 0.957
#> 2 x3 ~ x1 0.537 0.000
#> 3 x3 ~ x2 0.376 0.000
#> 4 x4 ~ x1 0.111 0.382
#> 5 x4 ~ x3 0.629 0.000
#> 6 x3 ~~ x3 0.874 0.000
#> 7 x4 ~~ x4 1.194 0.000
#> 8 x1 ~~ x1 0.933 0.000
#> 9 x2 ~~ x2 1.017 0.000
m <- matrix(c("x1", NA, NA,
NA, "x3", "x4",
"x2", NA, NA), byrow = TRUE, 3, 3)
p_pa <- semPlot::semPaths(fit_pa, whatLabels="est",
style = "ram",
nCharNodes = 0, nCharEdges = 0,
layout = m)
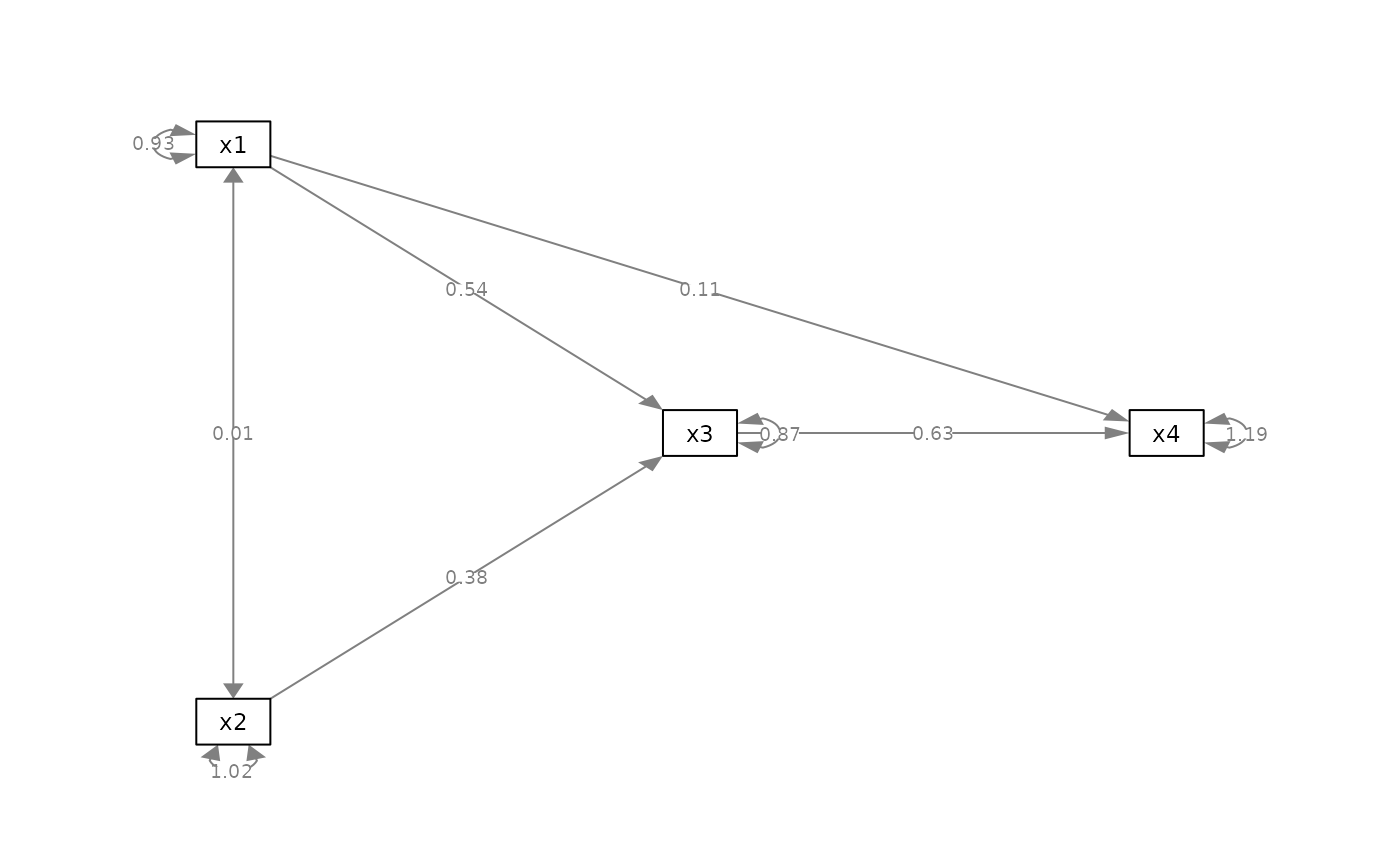 my_color_vector <- c("x2 ~~ x1" = "red",
"x4 ~ x1" = "blue")
p_pa2v <- set_edge_color(p_pa, my_color_vector)
plot(p_pa2v)
my_color_vector <- c("x2 ~~ x1" = "red",
"x4 ~ x1" = "blue")
p_pa2v <- set_edge_color(p_pa, my_color_vector)
plot(p_pa2v)
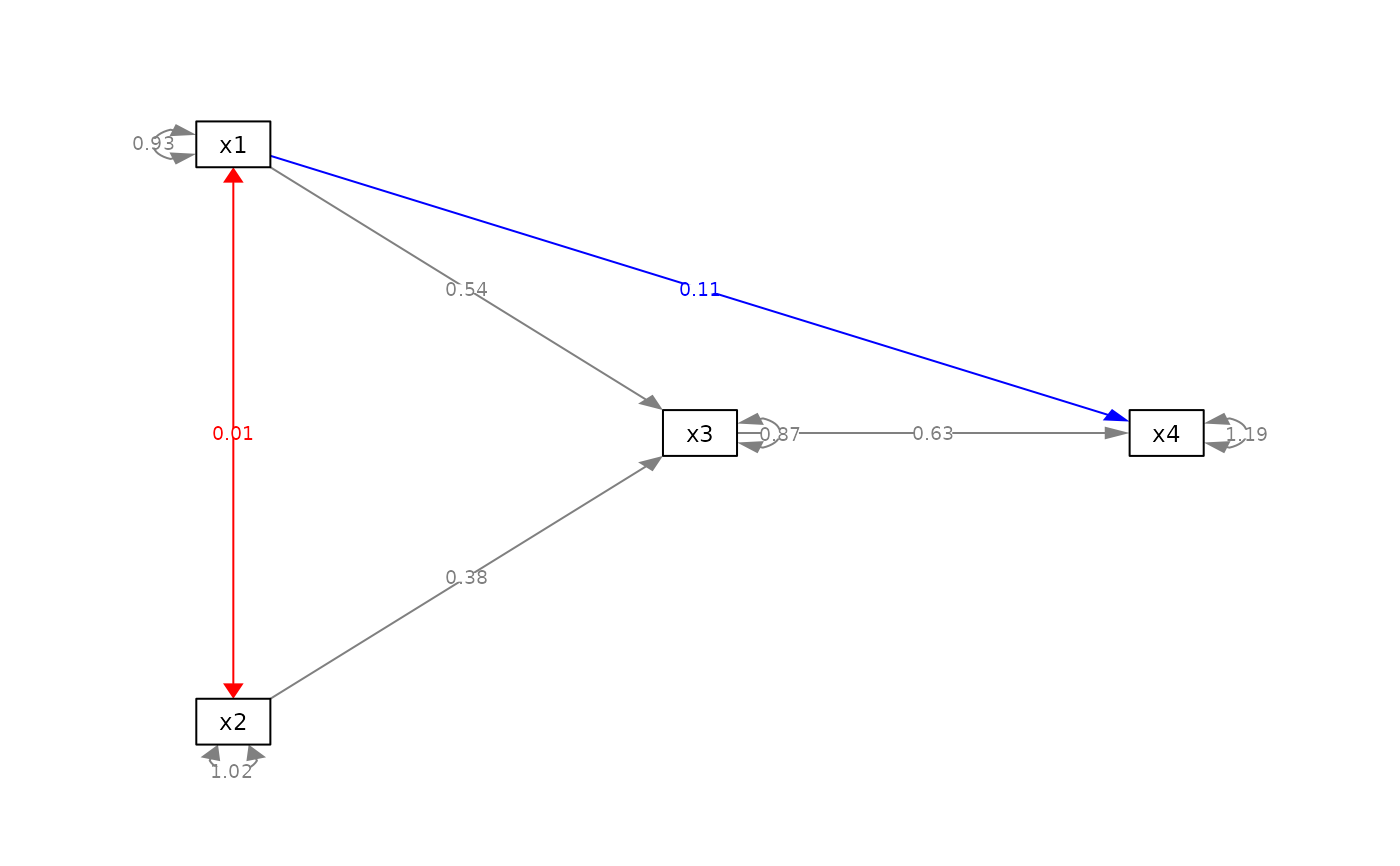 my_color_list <- list(list(from = "x1", to = "x2", new_color = "red"),
list(from = "x1", to = "x4", new_color = "blue"))
p_pa2l <- set_edge_color(p_pa, my_color_list)
plot(p_pa2l)
my_color_list <- list(list(from = "x1", to = "x2", new_color = "red"),
list(from = "x1", to = "x4", new_color = "blue"))
p_pa2l <- set_edge_color(p_pa, my_color_list)
plot(p_pa2l)
